
EU-LIFE Community Meeting 2024
It was a pleasure to be the host of the 11th EU-LIFE Community Meeting at CeMM in Vienna. Giulio Superti-Furga, Anita Ender and several CeMM representatives welcomed Marta Agostinho and the EU-LIFE office team, and the Board of Directors and Working Group members of the 15 EU-LIFE partner institutes.
The official EU-LIFE Community Meeting took place on Thursday, 25.4.2024, with more than 100 participants, contributing to 9 different Working Groups (Strategy, Core Facilities, Gender Equality and Diversity, Grants and Funding, IT, Human Resources, Training and Recruitment, Science Communications, and Tech Transfer). 7 out of 9 WG have already…
Read more
2. Place of Houska-Prize 2024 for Giulio Superti-Furga and his team at CeMM
Congratulations to Giulio Superti-Furga and his team for having been awarded the 2. Houska Prize 2024 of the B&C Private Foundation in the highly competitive category ‘University Research’.
The Houska Prize Jury recognized the accomplishment of having identified Feeblin as a new therapeutic approach for autoimmune diseases and for the effort to translate this finding by developing a new drug in cooperation with the domestic business partner and CeMM spin-off company Solgate GmbH that was founded in 2020.
Giulio Superti-Furga and his team at CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences have created a…
Read more
Ready and Vigilant: Immune Cells on Standby
When pathogens invade the body, the immune system must react immediately to prevent or contain an infection. But how do our defence cells stay ready when no attacker is in sight? Scientists from Vienna have found a surprising explanation: They are constantly stimulated by healthy tissue. This keeps them active and ready to respond to pathogens. Based on this insight, future medications could be devised to selectively enhance our immune system’s attention. The study has been published in the journal Nature Immunology (DOI: 10.1038/s41590-024-01804-1).
Communication is crucial in immune defence. When a virus infects a cell, the cell releases…
Read more
A shortcut for drug discovery: Novel method predicts on a large scale how small molecules interact with proteins
For most human proteins, there are no small molecules known to bind them chemically (so called “ligands”). Ligands frequently represent important starting points for drug development but this knowledge gap critically hampers the development of novel medicines. Researchers at CeMM, in a collaboration with Pfizer, have now leveraged and scaled a method to measure the binding activity of hundreds of small molecules against thousands of human proteins. This large-scale study revealed tens of thousands of ligand-protein interactions that can now be explored for the development of chemical tools and therapeutics. Moreover, powered by machine…
Read more
CeMM Research Report 2023: On the path to utopia
We would like to present you the CeMM Research Report 2023 dedicated to: on the path to Utopia in research. It represents our endeavour to embrace the joy of dreaming and visualizing the future.
What does the future hold? We have embarked on a journey to envision the future, going beyond traditional planning to truly 'dream it up'. Inspired by the EU-LIFE Utopia Conference in Lisbon in 2023, we posed this question to the CeMM community, inviting anyone interested to translate their vision of CeMM's various aspects and spaces into graphics for the 2023 Research Report. Several accepted the challenge. Principal Investigator André Rendeiro, who…
Read more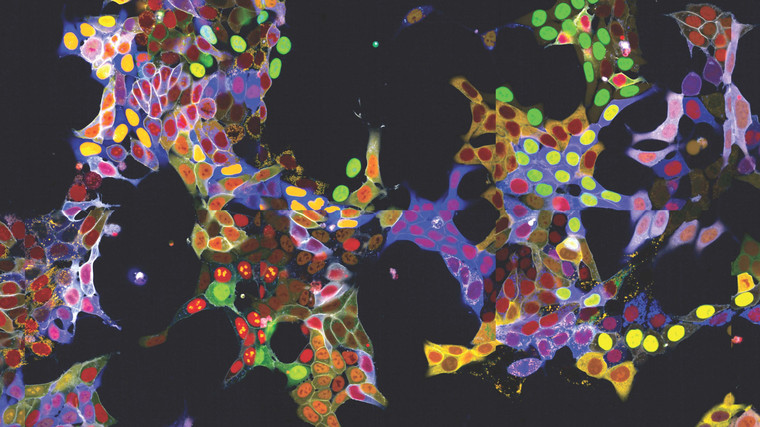
Light Show in Living Cells: New method allows simultaneous fluorescent labelling of many proteins
Observing proteins precisely within cells is extremely important for many branches of research but has been a significant technical challenge - especially in living cells, as the required fluorescent labelling had to be individually attached to each protein. The research group led by Stefan Kubicek at CeMM has now overcome this hurdle: With a method called "vpCells," it is possible to label many proteins simultaneously, using five different fluorescent colours. This automated high-throughput approach, aided by AI-assisted image recognition, opens up entirely new applications in various disciplines, from fundamental cell biology to drug…
Read more
ERC Advanced Grant for CeMM Adjunct PI Nuno Maulide
We congratulate Nuno Maulide, CeMM Adjunct PI and Professor at the University of Vienna, for receiving the ERC Advanced Grant - which makes him one of the very few people in Europe who have won all the European Research Council grants in a row (ERC Starting Grant in 2011, ERC Consolidator Grant in 2016, ERC Proof of Concept Grant in 2018 and 2023).
In his new ERC project "C-HANCE" hosted by the University of Vienna, Nuno Maulide explores the reactivity of positively charged carbon atoms (known as carbocations). Although many reactions involving carbocations are fundamental to organic chemistry - parts of which are even taught in basic…
Read more
EU-LIFE Charter of Independent Life Science Research Institutes
CeMM is a proud member of the EU-LIFE alliance since the beginning, and Giulio Superti-Furga, Scientific Director of CeMM and the Ri.MED Foundation, is the current Chair of EU-LIFE. The 15 EU-LIFE member institutes are leading and renowned life science research centers in Europe, and are profiting from the exchange of best practices, having common principles of research excellence, and contributing to European research policy. The different activities of the alliance are carried out by the Board of Directors and dedicated working groups, formed by experts and main representatives from the member institutions. They are supported by Marta…
Read more
New Insights Could Improve Treatment of Liver Fibrosis
Repeated or chronic liver injury, for example by viral hepatitis or alcohol consumption, triggers a complicated molecular process of scaring called liver fibrosis. Researchers at CeMM and MedUni Vienna have now succeeded in better understanding this process in a new study by examining gene activities at various stages of the liver disease. Their findings could contribute to a first-time therapy for fibrosis.
The liver is not only the largest internal organ but also vital for human life as a metabolic center. It also possesses remarkable self-healing powers: even when large portions are removed, such as during surgery, they quickly regenerate…
Read more
FWF "Emerging Fields" Grant for Brain Resilience
We congratulate the Consortium of the newly funded "Brain Resilience" grant! CeMM PI Christoph Bock is part of a team of world-renowned scientists that received one of the first "Emerging Fields" grants of the Austrian Science Fund (FWF). Over five years, the research groups coordinated by Igor Adameyko, MedUni Vienna will receive up to 6 million Euro to investigate brain resilience against disease.
The brain of mammals is formed through highly complex developmental processes, controlled by thousands of genes and their interaction with the prenatal environment. Mutations in the underlying genes can predispose individuals to various…
Read more